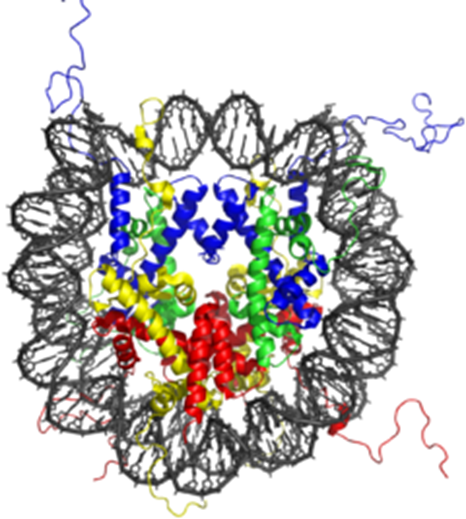
Nucleosomes are core particles involved in DNA packaging in eukaryotic cells. They affect gene expression levels by regulating access of DNA to transcription factors and DNA binding proteins. Nucleosome modeling is not static and chaotic. The link between nucleosome dynamics and gene expression modulation still remains a mystery, driving Life Scientists to design clever nucleosome-based in vitro assays such as those we’ll be reviewing here.

Research solutions enabling a better understanding of nucleosome dynamics and chromatin modeling are crucial nowadays.
HeLa nucleosomes, biotinylated 601 sequence DNA and recombinant Human Histone octamers are commercially available for numerous applications, for research and Drug discovery screening applications:
- Enzymatic assays (ex. HAT assay)
- Enzymatic modification assays, (ex. methylation)
- Chromatin reconstitution experiments
- Pull-down of nuclesomes, especially useful in nucleosome binding assays
- ChIP assays
- Transcription and histone modification assays
Let’s take a closer look at the most popular nucleosome-based assays that researchers mention during our daily interaction in the field of epigenetic studies and drug discovery screenings. It’s interesting to note that the robustness and purity of nucleosome preparations are frequently highlighted.
Why develop assays related to Nucleosomes, Chromatin modeling and Gene expression?
By forming the fundamental repeating core units of eukaryotic chromatin (yeast, worms, flies, humans…), nucleosomes pack the large eukaryotic genomes into the nucleus ensuring appropriate access to it. They consist of a 147bp DNA sequence bound with a Histone octamer and are connected together via a 80 bp long DNA linker (2 copies each of Histones H2A, H2B, H3, and H4 ).
Nucleosome modeling is a precise regulatory control element modulating gene expression patterns during cell life. Nucleosomes are thought to carry epigenetically inherited information through post-translational modifications of their core Histone components (by HATs, HDACs…). Such modifications impact several key biological processes such as DNA repair, replication, differentiation, and apoptosis…
The dynamic distribution of nucleosomes (chromatin remodeling) alongside the DNA is a non-random and strictly controlled process throughout the various stages of the cell cycle. This fine-tuned “nucleosome landscape” brings a higher level of gene expression regulation by enabling (or not, depending on the level of DNA condensation) “genomic DNA- transcription factor” interaction and DNA binding protein activity. (1 – 5)
Nucleosome composition and architecture together with chromatin remodeling are enzyme-driven. Understanding DNA condensation, nucleosome landscape dynamics and epigenetic events is crucial if one wants to move forward in gene expression studies, drug discovery and personalized medicine.
But what are the most popular nucleosome research tools available for Histone Modification Assays and screening of small molecular inhibitors for drug discovery and HTS applications?
Research tool #1 – Biotinylated Nucleosome Assembly 601 Sequence DNA – a simple way to focus on DNA / Histomer octamer binding
Biotinylated Nucleosome Assembly 601 Sequence DNA is a 186 base-pair double-stranded DNA fragment that was identified by Lowary and Widom (1998) using the SELEX method. The 601 sequence DNA has high affinity for Histone octamers.
This 601 sequence DNA has a biotin group on the 5’ end which makes it ideal for use in in vitro nucleosome assembly using recombinant or purified octamers (see below), nucleosome binding assays and pull-down experiments.
The biotinylated Nucleosome Assembly Sequence 601 DNA is very often available in lyophilyzed format for convenient shipment and storage. After resuspending, 601 DNA aliquots must be stored at -80°C.
Research tool #2 – Purified Chicken Oligonucleosomes – the cheapest alternative to nucleosome assays

Purified Chicken Oligonucleosomes are extracted from chicken erythrocytes. They are predominantly composed of Histone hexamers, septamers and octamers.
They remain, in any event, very affordable nucleosome alternatives for Life Scientists seeking to develop enzyme assays such as acetylation, methylation or phosphorylation, chromatin binding assays, or for use as a positive control in Western blotting. Purified chicken oligonucleosomes usually have a concentration of 2.5 mg/ml (DNA + protein) in 10 mM Tris-HCl pH 8.0, 1 mM EDTA and 10% glycerol.
Chicken Oligonucleosomes are stable (for six months at -80°C). For best results, aliquot and avoid multiple freeze/thaws.
Research tool #3 – Purified HeLa Poly- or Mono-nucleosomes – highly purified and well-validated Human nucleosomes for enzyme assays
These Human nucleosomes are purified from HeLa cells at differing concentrations (often 6.0 mg/ml and 5.5 mg for poly- and mono-nucleosome respectively) in HEPES buffers to be used as substrates for enzyme assays.

Purified HeLa Polynucleosomes (mostly di-, tri- and tetramers) and Mononucleosomes are ideal for performing enzyme assays (Acetylases, Methylases, Kinases / Phosphatases, DUBs…), chromatin binding assays, or for use as a positive control in Western blotting.
Notably, nucleosomes from EpiCypher are purified with a unique and proprietary process ensuring the production of high quality products with minimal lot-to-lot variations, reducing false positive or negative results.
Research tool #4 – Human recombinant Mononucleosomes – biotinylated or without free DNA for high quality enzyme testings
Recombinant Mononucleosomes with two each of histones H2A, H2B, H3 and H4* are assembled from recombinant Histones expressed in E. coli and wrapped by 147 base pairs of 601 sequence DNA.
Available at a concentration of 0.5 mg/ml in Tris-HCl buffer, recombinant Human mononucleosomes have 3 key characteristics:
- They are highly purified
- They do not contain free DNA
- They do not present any post-translational histone modifications
These unique characteristics make Recombinant Mononucleosomes ideal substrates for Life Scientists running enzyme activity and screening assays and looking for robust experiments and reproducible data.
Recombinant Mononucleosomes are suitable for nucleosome binding experiments too.
*Accession numbers for recombinant Histones used in EpiCypher’s recombinant Mononucleosomes: H2A-P06897; H2B-P02281; H3-Q92133; H4-P62799.
Recently, a biotinylated format of Human recombinant Mononucleosomes has become available. In this format, a 40 base pair spacer sequence with a biotin group on the 5’ end of the DNA has been added. These Human recombinant biotinylated Mononucleosomes have been requested by researchers running nucleosome binding assays and pull-down experiments.
Research tool #5 – Human recombinant Histone Octamer – access a nucleosome protein component for 100% controlled chromatin experimental design
Recombinant Human Histone Octamers are recombinant H2A, H2B, H3 and H4 Histones expressed in E. coli (accession numbers used: H2A-P0C0S8; H2B-P62807; H3-P68431; H4-P62805) and purified by gel filtration. These highly purified recombinant Human Histone octamers (1 mg/ml) in Tris buffer can be used for chromatin reconstitution experiments, or as substrates for histone modifying enzymes.

Sources:
1/ McDonald D, “The nucleosome hypothesis: An alternative string theory” (2005) Nature Milestones: Gene Expression. DOI : 10.1038/nrm1798.
2/ Kornberg RD “Chromatin structure: a repeating unit of histones and DNA” (1974) Science, 184 – 4139, 868–71. DOI :10.1126/science.184.4139.868.
3/ Lorch Y et al. “nucleosomes inhibit the initiation of transcription but allow chain elongation” (1987) Cell, 49, 203–10. DOI:10.1016/0092-8674(87)90561-7.
4/ Han M and Grunstein M “Nucleosome loss activates yeast downstream promoters in vivo”(1988) Cell, 55, 1137–45. DOI: 10.1016/0092-8674(88)90258-9.
5/ Bunnik EM “DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum” (2014) BMC Genomics, 15-347. DOI: 10.1186/1471-2164-15-347
Want to know more about Nucleosome components, recombinant or purified nucleosomes for enzyme screenings, binding assays and chromatin modeling?
Nowadays, high quality nucleosome-related research tools are available to ensure accurate experimental results.
These research tools (together with primary antibodies, cell-based assays…) bring optimal solutions to Life Scientists for deciphering chromatin biology and epigenetics. This is also true for bioactive and pure Small Molecules (ex. HDAC Effectors (Tubastatin, MS-275, Trichostatin A, Apicidin, Panobinostat…), HAT Effectors (C646, Garcinol…) and SIRT Modulators (EX-527, BML-278, Resveratrol and its natural analog, Piceatannol, Sirtinol…).
If you want to know more about these solutions and get in touch with experts, you might like to sign up for thematic tebu-bio newsletters, or contact them directly.
[contact-form to=’myprotein@tebu-bio.com’ subject=’Nucleosome-based research tools’][contact-field label=’Name’ type=’name’ required=’1’/][contact-field label=’Email’ type=’email’ required=’1’/][contact-field label=’Comment’ type=’textarea’ required=’1’/][/contact-form]