To dilute or not to dilute biological samples? And if yes, how much? That’s the question!
Following our post on whether samples should be pooled or not when performing studies for Biomarker discovery, it’s now time to discuss sample dilution (yes or no) and dilution factor (how much) when performing proteomic analysis of Biomarkers.
Dilution of samples – yes or no?
For proteomic analysis, usually the answer is yes.

But what happens when you have a very, very limited amount of sample, and moreover, your samples are really precious (e.g. clinical samples)? The answer is still yes. Positively.
Let’s stop for one second and think why we want to analyse precious, limited samples. We want to analyse them because we want to find relevant biomarkers. We don’t want to get artifacts due to different molecules present in our sample. Let’s take plasma. The amount of proteins found there (which are general proteins, and not biomarkers) can hinder the interpretation of our results, and cause false positives due to background. More if the plasma is hemolysed.
Serum is pretty much the same. I’m thinking about albumin, but also about auto-antibodies, and a lot of debris that can also be detected in the wavelengths currently used to calculate Abs values, and therefore, used to determine whether a change in the health status links to a change in a given biomarker.

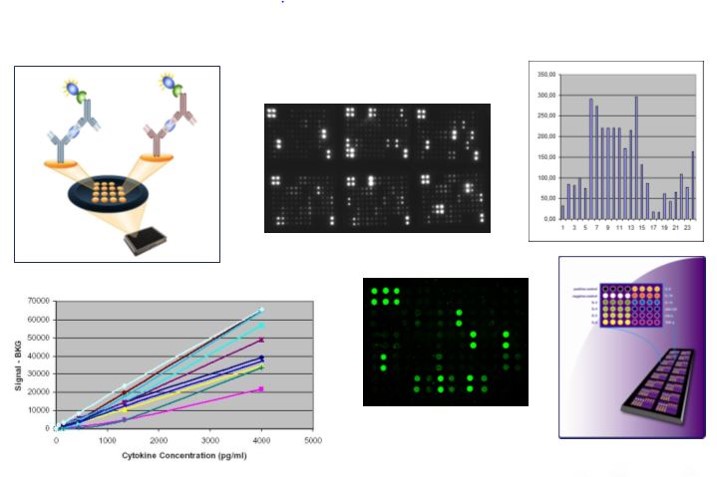
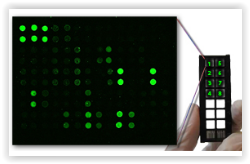
Using optical technologies, such as antibody arrays or optical-based multiplex ELISAs is an advantage, as any background will be clearly visible in the scanner or optical instrument (and we can perform additional washes to remove – at least partially – the background signal). There is certainly a problem when using classical ELISAs or bead-based technologies, as in this case, we only get a value, and cannot really “see” how the experiment worked.
We will discuss on the “how much” in another post. But please remember…you should always dilute your samples, at least a 1:2 fold. Quite frankly, publications using undiluted samples should either show validation of results using different technologies, or quite honestly, one cannot be certain whether the values obtained come from a real biomarker detection or just background noise.
This can be a topic of debate… feel free to leave your comment!