In a previous post, we discussed kinome studies in the tumour microenvironment (TME). We described some solutions to study known markers, but we did not look at those cases in which the biomarkers associated to the kinome are unkown, and therefore some exploration is needed.
Case # 1 – when you know the way
When you have a rough idea of the pathway involved in your experimental model, you normally study it. As simple as that. In my previous post, I described different techniques that can be used when you know which kinase is relevant to your biological process. However, in some cases, you may need to study the different positions that can be phosphorylated in your kinase, or going one step beyond, study other upstream or downstream signaling biomarkers.
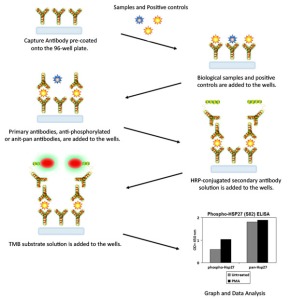
If this is the case, then phospho-ELISAs may be of help. They also allow you to get higher throughput than single antibodies. There are phospho-ELISAs for given kinases, in order to study different phosphorylations. For example, one may be interested in studying phosphorylated and non-phosphorylated forms of EGFR, which is an important biomarker in lung cancer.
Also for lung cancer, one may be interested in studying only the phosphorylation of S473 in Akt, in a large number of samples (e.g. when testing different drug candidates to see their effect).
In some gynecological cancers, one may be interested to study phosphorylation of the Tyr residues in JAK1 to unravel the immune regulation mechanisms in cancer progression.
Soon, new ELISAs will be released to study several kinases, simultaneously, in a given pathway (e.g. Akt/GSKb/mTOR, ERK/JNK/p38a).

Case #2 – when you don’t know the way
Let’s suppose that you have read a lot of publications and have a rough idea about what pathways cooperate to the genesis and progression of cancer in your experimental model. Still, you want to know a bit more, and discover new pathways… or explain some drug resistances that you are finding in your patients. Another possibility is that you have a drug candidate, and want to study its mechanism of action.
In this case, you want to do a broad profiling on the pathways that are relevant. You can do this either by studying the expression at the mRNA level, or at the protein level.
At the mRNA level, you will be able to study those genes which have been described as relevant for different types of cancer, or involved in drug resistance. This will provide information at the transcriptional level, indicating whether a gene is up- or down-regulated.

If you need to go a step further to know not only the expression levels, but also what post-translational modifications have taken place (i.e. phosphorylations), then you need to go to the protein level. You may choose between studying “popular” pathways (e.g. AMPK, mTOR, Jak/STAT, etc), go for the cancer pathways, or do a whole profiling for more than 1,300 biomarkers.
Alternatively, you may wish to study the phosphorylation of receptors such as EGFR and RTKs. These receptors have a key role in the development of some cancers, as well as drug resistance (e.g. EGFR in lung cancer). For these arrays, detection is based on a sandwich technique (capture-detection), rather than single antibody detection (as it is the case for arrays studying kinases).
So… ready to explore?