Following on from my previous post in which I started to introduce a series of techniques to investigate the impact of miRNA expression on cell fate, let’s take a look at the first one: miRNA detection
miRNA databases
There are many miRNA databases available such as miRBase: the microRNA Database . It’s very complete, very intuitive, and you can find information on sequences, targets and tissular location of miRNAs to name but a few.
miRNA detection by qPCR
This is the most classical way to detect miRNAs. You must have reliable (validated) primers and choose a reliable house keeping short miRNA or other non-coding RNA sequence as a normalization control.
Good practice is to choose 2 or 3 housekeeping genes to circumvent situations when even housekeeping can be altered under treatment. As a matter of fact, there is an interesting discussion topic on how to normalize RT-qPCR based micro-RNA expression data in humans using cel-miR-39 in the microRNA research LinkedIn group, where most participants agree to include 2-3 different miRNA as housekeeping for data normalization.
Here are several miRNA primers sets that you might find of interest.
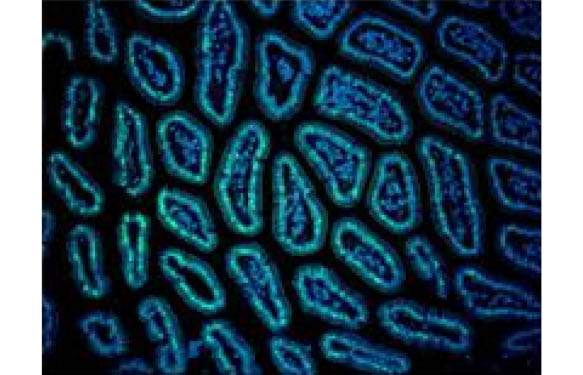
Labelled aptamers for microRNA detection
Aptamers are small oligonucleotides with high affinity for proteins, RNA or DNA. Fluorescent aptamers can be used as probes to detect miRNA in toto (immunohistochemistry) or at individual cell level.
Profiling miRNAs
Profiling plates offer a panoramic view of the whole miRNA signature of cells or tissues in one single experiment. I’ll come back to this point in more detail in a later post in this series!
Questions, anyone? Let me know!