Epigenetic modifications target DNA and Histone proteins.
One of the roles of Epigenetics is to drive cellular differentiation from totipotent Stem cells to fully differentiated cell types, by regulating gene expression through patterns of modifications that differ according to the cell type and the cell status. This means that the patterns may significantly differ between stem cells, germ cells, and differentiated cells.
It has been found that cancer cells often differ in regard to specific Epigenetic modification sites too. Thus, epigenetic enzymes are considered as potential pharmaceutical drug targets or biomarkers, and quite some effort is made to define drugable targets.
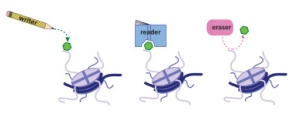
In this post, I’d like to take a look at Histone modifications, and especially histone modifying enzmes (writers). Later on, in further posts I’ll be focusing on enzymes which recognize specific modifications (readers), and those that erase modification from histones (erasers).
Which kind of epigenetic histone modifications are known?
#1- Lysine and Arginine Methylation
While Lysine can be mono-, di-, or trimethylated, Arginine can be mono-methylated on one of the amino groups of the side chain, or di-methylated either on one of the amino groups (asymmetric dimethlyarginine) or mono-methylated on both amino groups (symmetric dimethlyarginine).
As lysine methylation events leave the charge of the lysine intact, and only add a very small number of atoms, the steric interactions are basically unaffected. Nevertheless, specific Reader domains on modifying enzymes (a topic of a future post) can differentiate between the site and the number of methylations; methylation pattern being linked to most of the known functions of histone modifications.
#2- Lysine Acetylation
Unlike methylation, acetylation of lysines has a significant effect on the amino acid because this modification neutralizes its positive charge. This leads to a reduction of the electrostatic attraction between the DNA backbone which is negatively charged and the histones (which upon acetylation have lost their positive charge). Finally this process results in a loosening of the chromatin structure, meaning the chromatin becomes more accessible and active transcription increases in these chromatin regions.
#3- Serine, Threonine or Tyrosine Phosphorylation
The exact implication of histone phosphorylation is not yet known, but generally an addition of a negatively charged phosphate can lead to distinct changes in protein structure, which might as well have a significant impact on chromatin structure and accessibility.
#4- Lysine Ubiquitination
Mono-Ubiquitination of different histones can either lead to increase or even silence transcription events.
#5- Poly ADP Ribosylation
Poly ADP Ribosylation of histones has been described as a mechanism to regulate histone methylation events.
#6- Other types of Histone modifications
This includes Sumoylation, Carbonylation, and Glycosylation. Their impacts on protein and cellular functions are not fully understood yet.
What’s new that I can use for my research?
When you start screening activities on new targets you might need to establish and optimize new assays in your laboratory.
To accelerate your assay development, BPS Biosciences has released a wide variety of HTS compatible ready-to-use assays to run inhibitor screenings for a number of epigenetic enzymes.
You might like to take a look at…
Methyltransferase activity assays, Acetyltransferase activity assays, Poly ADP Ribose Polymerase activity assays
Research tools reviewed in this post can definitely help you in your epigenetic research – in drug discovery activities and screening for inhibitors of epigenetic target enzymes and in academic and fundamental research as well.
Don’t hesitate to ask your questions or share your comments below!