Lung cancer is one of the most common malignancies, and the leading cause of cancer-related fatality. Current diagnostic practices for common cancers rely heavily on imaging technologies. These methods are quite accurate, but still have a probability of having false-positive findings. Also, there is a substantial need for non-invasive ways to test whether the nodules are benign or malignant.
Blood-based biomarkers have potential in cancer screening, and their role could extend further from general population risk assessment to treatment response evaluation and recurrence monitoring. However, despite much research effort, biomarkers able to predict disease onset and evolution are not always easy to find, or distinctive enough.
Protein markers currently in clinical use have limitations with respect to their use for screening owing to low sensitivity and specificity in early stages, and inability to distinguish aggressive from indolent tumors. Also, lung cancer lacks established biomarkers with demonstrated clinical utility in a screening setting. Thus, there is a need for biomarkers with the required sensitivity and specificity for the detection of frequently occurring cancer types.
Classically, Proteomics technologies seek to identify and sequence annotate the proteome by LC-MS/MS analyses of peptides derived through proteolytic processing of the parent proteome. Thousands of proteins have been identified from human serum as a result, which has allowed the development of more precise tools, i.e. antibody arrays. These antibody arrays include only those markers that may be relevant and have been previously described. Still, some research is still needed to validate them in profiling & validation projects for, say, secretome or kinome markers.
Still, there are fields where our knowledge can definitely advance more, e.g. functional annotation. This is crucial, as the landscape of protein conformations is highly variable, and each conformation may contribute to its own functional activity. Sequence annotation alone cannot capture this vital information, so new strategies are necessary. The challenge has been to overcome the analytical bias toward the most abundant proteins, and the complexities of mining the data to a manageable number of biomarker proteins that can be analyzed in more depth.
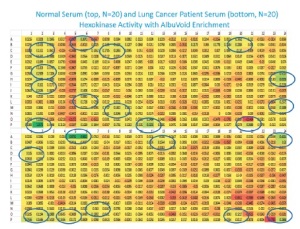
A series of new tools allowing to combine array technologies with 2D electrophoresis, without compromising functionality, have been developed. A review on these technologies and how they can be used for biomarker discovery in different types of cancer has been recently published in GEN, where they found a link between hexokinase activity and lung cancer.
If you would like to receive a copy of this review, get in touch with us!