Antibody pairs have been used for years to detect biomarkers in biological samples. They are a convenient and price-wise alternative to ready-to-use ELISAs, especially when they are used on a routine basis (for sporadic use, ELISA kits may be a better choice).
These classical tools are now having a revamp in some research areas, not only the “classical” ELISA use, but also IP-WB and even protein-protein interaction studies.
#1 – ELISA for biomarker quantification

The trend nowadays is to run profiling projects, allowing to discover new biomarkers. This may mean that, for some of these new markers, ready-to-use ELISAs are not available.
Developing a new ELISA can be a lengthy and costly project. This is one of the reasons why some laboratories use other immuno-assays (e.g. WB, IHC, IF) to validate the results obtained used profiling arrays. These techniques are very reliable (particularly if you use validated antibodies), but they are not quantitative.
You might not be aware, but there are thousands of validated, matched antibody pairs for really exotic targets (not the usual cytokines). Here are my favourite ones for ELISA applications:
- BLNK (Human) Matched Antibody Pair -> BLNK bridges B cell receptor-associated kinase activation with downstream signaling pathways, thereby affecting various biological functions (2).
- REG1A (Human) Matched Antibody Pair -> REG1A is associated with islet cell regeneration and diabetogenesis and may be involved in pancreatic lithogenesis (3).
# 2 – IP-WB for biomarker discovery
IP-WB is a powerful tool to enrich specific proteins that may be present in low levels in our samples. It is also useful to enrich specific-protein modifications (e.g. phosphorylation, ubiquitination) in heterogeneous samples (e.g. cell lysates). In brief, the IP antibody recognises the specific protein or modification in its native or semi-native format. After IP, a WB can be used for analysis.
- STYK1 (Human) Matched Antibody Pair -> STYK1 plays important roles in diverse cellular and developmental processes, such as cell proliferation, differentiation, and survival (4).
- ICAM1 (Human) Matched Antibody Pair -> ICAM1 is a cell surface glycoprotein which is typically expressed on endothelial cells and cells of the immune system. It binds to integrins of type CD11a / CD18, or CD11b / CD18 and is also exploited by Rhinovirus as a receptor (5).
# 3 – Protein-protein interactions for biomarker discovery
Some new therapies are aimed not at a target per se, but at disrupting its interaction with other targets. Protein-protein interactions (PPI) play a central role in the regulation of biological processes. However, before this process can be considered as “druggable”, much research is needed to understand the nature and behaviour of these PPIs. In situ Proximity Ligation Assays (PLA) can certainly be of help in this area of research.
Use of PLA antibody pairs presents one major advantage, as they allow to see the PPIs directly in cells (even in fresh cells, no need to fix). This means that we will not only see the proteins interacting, but most importantly, their differential localisation depending on the treatment / stimulus applied to the cells. Functional exploration of a disease network with interlinked pathways via PPIs (i.e. interactome) can be used to discover novel biomarkers (6).
Here are my favourite antibody pairs to study PPIs:
- CDKN1A & CDKN1B Antibody Pair for PLA -> some genetic variations in CDKN1A and CDKN1B may be involved in some cancer types, and this may affect their interaction (7). Also, Tbx2-mediated regulation of these markers may play a crucial role in cell cycle regulation during morphogenesis (8).
- ERBB3 & ERBB2 Antibody Pair for PLA ->ERBB3 and ERBB2 interactions have been described to be associated with mAb therapy resistance in some cancer types (9).
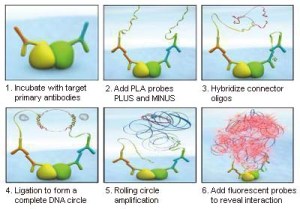
In situ Proximity Ligation Assay (PLA) – how it works!
In conclusion, whether you are using the “classical” techniques such as ELISA or IP-WB, or want to unravel new mechanisms in the interactome, antibody pairs are back to help you!
Any questions? Just leave your comments below!
References
1.- Yang T (2013) PEGylation – Successful Approach for Therapeutic Protein Conjugation. Mod Chem appl 1:e112. doi: 10.4172/2329-6798.1000e112.
2.- http://www.ncbi.nlm.nih.gov/gene/29760.
3.- http://www.ncbi.nlm.nih.gov/gene/5967.
4.- http://www.ncbi.nlm.nih.gov/gene/55359.
5.- http://www.ncbi.nlm.nih.gov/gene/3383.
6.- Chia-Hung Liu, Tzu-Chi Chen et al. Mol Cell Proteomics 2013 12: 1335–1349. doi:10.1074/mcp.O112.020404.
7.- Hongxia Ma, Guangfu Jin. Int. J. Cancer: 119, 2173–2178 (2006). doi:10.1002/ijc.22094.
8.- Lüdtke TH-W, Farin HF, Rudat C, Schuster-Gossler K, Petry M, et al. (2013). PLoS Genet 9(1): e1003189. doi:10.1371/journal.pgen.1003189.
9.- Xiaoping Huang et al. Cancer Res February 1, 2010 70:1204–1214; doi:10.1158/0008-5472.CAN-09-3321.




One Response
Very informative post related to bioinformatics. It is great help for my new project .By reading this article, I learn some important things that I need to improve. I continuously check this site for regular updates in field. Thanks for putting top notch content in article. I would like to be here again to find another masterpiece article.