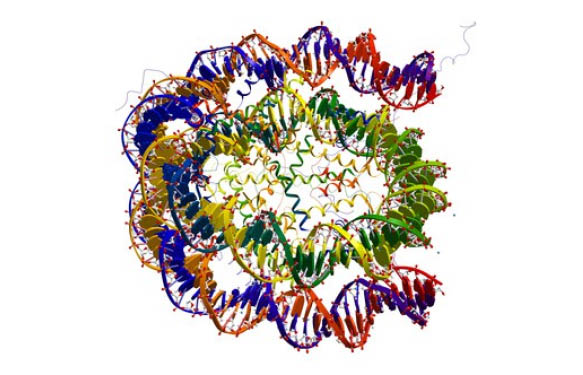
Nucleosomes are basis units of DNA packaging in eukaryotic cells. A core particle (mono-nucleosome) consists of a segment of DNA called core DNA (147 bp in length) wound around a histone octamer (Fig 1.). Histone octamers are made up of 2 copies of the the core histones H2A, H2B, H3, and H4. These mono-nucleosomes are connected by linker 80bp-long DNA. H1 histone, the so-called linker histone, binds to the linker DNA close to the entry and exit of the core DNA and is involved in chromatin compaction (Fig 1.).
Histone and DNA modifications regulate gene expression

The eukaryotic genome is organized in chromatin. In order to regulate processes such as transcription, DNA replication and DNA repair, the chromatin structure has to be modulated in order to change the condensation level. Different enzymatic (epigenetic) modifications are correlated to either gene activation or gene silencing.
While methylation of DNA and histones causes nuclesomes to pack tightly together (thus hindering e.g. transcription factors to bind the DNA and to cause gene expression ), histone acetylation for instance cause loosening of the packing of nucleosomes (thus allowing access of factors and gene expression (Fig. 2)).
Nucleosomes are substrates for enzyme assays and inhibitor screenings
To measure the activity of histone modifying enzymes and screen possible inhibitors, peptides derived from specific histone sequences are often used in in vitro assays. These peptides can either be “blank” for methyltransferase reactions, or specifically modified for instance for demethylase or deacetylase reactions. Nevertheless, for many chromatin modifying enzymes, nucleosomes are a more physiologically relevant substrate than these peptides.
Furthermore, the quality, and especially the purity of nucleosome preparations (native or put together from recombinant histones) is crucial to get reliable activity and inhibition results. By using highly purified nucleosomes, screeners can significantly reduce false positive and false negative hits. Especially contaminating free DNA or un-complexed histone units can lead to imprecise results.
EpiCypher have optimized their protocols for preparing different nucleosome preparations that meet the standards expected by researchers when using nucleosome-based assays. These high purity nucleosome preps show superior characteristics vs. competitors’ preps. In Fig. 3, the absence of free DNA in EpiCypher’s recombinant nucleosomes is clearly seen (as compared with nucleosomes contaminated with free DNA produced by another source).

Regarding recombinant nucleosomes, comparative analysis of EpiCypher’s production vs. other sources clearly shows significant differences on the protein level. In addition to His-tags and/or biotins added to the final construct, SDS-PAGE analysis displays multiple protein bands; the 4 histone types being not equally represented.

My selection of nucleosome preparations
For your in vitro assays related to inhibitor screening, epigenetics studies and chromatin reconstitution, I would recommend you to use the following preps:
Mono-nucleosomes here are assembled from recombinant human histones expressed in E. coli (two each of histones H2A, H2B, H3 and H4 wrapped by 147 base pairs of 601 positioning sequence DNA). There is a 5’ biotin-TEG group on the DNA which makes them ideal for use in nucleosome binding assays and pull-down experiments.
These human mononucleosomes are purified from HeLa cells and consist of the histone octamer (two each of the four core histones, H2A, H2B, H3 and H4) wrapped by 147 base pairs of DNA.
Human polynucleosomes are purified from HeLa cells. HeLa Polynucleosomes are predominantly trimers, with some dimers and tetramers
Oligonucleosomes are purified from chicken erythrocytes. Chicken Oligoucleosomes are predominantly hexamers, septamers and octamers.
These human histone octamers (two each of histones H2A, H2B, H3 and H4) are made from recombinant histones expressed in E. coli.
Nucleosome Assembly 601 Sequence DNA, Biotinylated is a 147 base-pair double-stranded DNA fragment which has high affinity for histone octamers and is useful for in vitro nucleosome assembly. There is a biotin group on the 5′ end of the fragment, which makes it ideal for use in nucleosome binding assays and pull-down experiments.
Looking for nucleosomes and related products?
Get in touch with me through the form below.
[contact-form to=’ali.el.baya@tebu-bio.com’ subject=’Keen to work with the best quality nucleosomes?’][contact-field label=’Nom’ type=’name’ required=’1’/][contact-field label=’Email’ type=’email’ required=’1’/][contact-field label=’Commentaire’ type=’textarea’ required=’1’/][/contact-form]