In a previous post on tumour microenvironment (TME) and glycosylation, we indicated that new tools for studies on glycosylation of proteins were being developed. We are happy to announce that they are now available!
 These new tools are directed at studying the glycosylation of proteins in the secretome and inflammasome. This is related to TME, but also to many other processes where factors such as cytokines, chemokines, adipokines, growth factors, angiogenic factors, proteases, soluble receptors and soluble adhesion molecules are relevant.
These new tools are directed at studying the glycosylation of proteins in the secretome and inflammasome. This is related to TME, but also to many other processes where factors such as cytokines, chemokines, adipokines, growth factors, angiogenic factors, proteases, soluble receptors and soluble adhesion molecules are relevant.
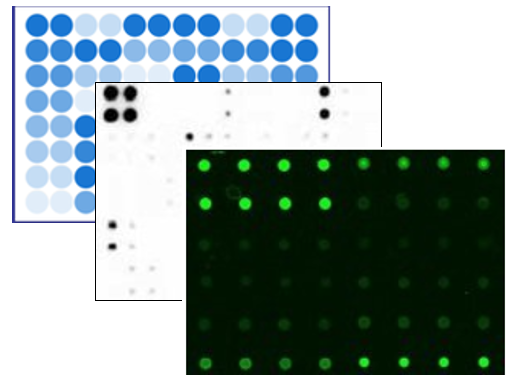
Compared to lectin-based arrays, these new arrays allow you to not only to study the glycosylation pattern, but also to identify which biomarker is glycosylated. Specifically, capture antibodies for 507 different secretome factors are printed onto glass slides, and the glycans on these capture antibodies are removed. After incubation with samples (serum, plasma, lysates) and washing to remove unbound proteins, five biotin-labeled lectins are incubated with the array to allow binding to the glycans from captured target proteins. Detection is done with a streptavidin-conjugated fluorescent dye and laser fluorescence scanning. The obtained signals are then compared to the array map to identify glycosylated proteins present in the samples.
These arrays are brand new… and open a completely new path for the discovery of glycosylation-related biomarkers in a wide variety of diseases.
Ready to explore? How do you think these could help your research?