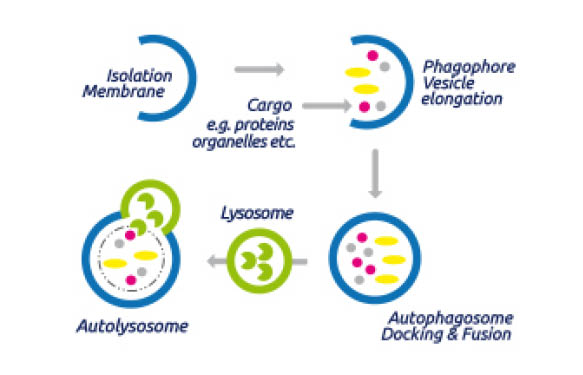
The term Autophagy was introduced by Christian de Duve during the Ciba Foundation Symposium on Lysosomes – which was held in London in February 1963. In 1974 he was honoured with the Nobel price in Physiology or Medicine for his pioneering research about peroxisomes and lysosomes.
In this post, I’d like to give you an overview of autophagy and its implication in cell biology, and tools to manipulate and detect autophagy in cells.
Autophagy (Autophagocytosis) describes the fundamental catabolic mechanism during which cells degrade dysfunctional and unnecessary cellular components. This process is driven by the action of lysosomes and promotes survival during starvation periods as the cellular energy level can thus be maintained.
During classical autophagy, autophagosomes are generates, organelles which are surrounded by double membranes (see Fig. 1) and which contain the cellular components to be degraded. The autophagosome formation is induced by class 3 phosphoinositide-3-kinase, Atg 6 (autophagy-related gene 6) and ubiquitin or ubiquitin-like modifications of the target proteins.
Subsequently, autophagosomes traffick through the cytosol of the cell and finally fuse with lysosomes to form an autolysome. After this fusion the cargo is degraded by lysosomal hydrolases.
Role of autophagy in cellular processes
Autophagy plays a crucial role in several basic cellular and pathological processes (Fig. 2).
As already indicated, nutrient starvation induces a high level of autophagy to maintain cellular energy levels. Furthermore, autophagy plays an important role in innate immunity as pathogens which invade cells (e.g. Mycobacterium tuberculosis) can be targeted for degradation with the same mechanism described above. As autophagy leads to the degradation of damaged organelles, it can also function as a repair mechanism. And finally the appearance of autophagosomes is associated with one of the mechanisms of programmed cell death (type II cell-death).
Role of autophagy in pathological processes
More and more findings indicate that alterations in the autophagic degradation process, especially mutations in the ATG genes (autophagy-related genes) can be linked to a number of neurodegenerative diseases, infectious diseases, cancers and other diseases (for a summary, see Jiang and Mizushima, Autophagy and human diseases, Cell Research 24:69–79, [2014]).
Thus, increasing interest in drug discovery and fundamental research is directed towards autophagy-modulating compounds.
Tools to manipulate autophagy
To allow easy access to all compounds showing an effect on autophagy, I’ve listed them here with their main features.
Chloroquine phosphate – an anti-malarial drug that inhibits autophagy in a variety of cell lines. Induces cell death in breast cancer cell lines and displays antitumor and antimetastatic activity in mouse models of breast cancer. Eliminates cancer stem cells via deregulation of Jak2 and DNMT1. Chloroquin phosphate displays synergy when combined with the Raf inhibitor, vemurafenib, in brain tumor4. Cell permeable and active in vivo.
C2-Dihydroceramide – usually used as a negative control for C2 Ceramidea. Can also function as an autophagy inducer.
Dorsomorphin – Potent, selective and ATP-competitive inhibitor of AMP-activated protein kinase (AMPK) (Ki = 109 nM). Also inhibits bone morphogenetic protein (BMP) type I re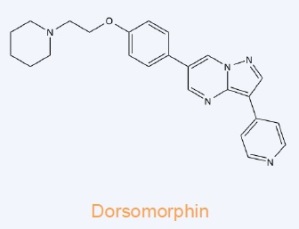 ceptors (ALK2, ALK3and ALK6). Induces autophagy in cancer cell lines via a mechanism independent of AMPK inhibition.
ceptors (ALK2, ALK3and ALK6). Induces autophagy in cancer cell lines via a mechanism independent of AMPK inhibition.
Hydroxychloroquine sulfate – anti-malarial and anti-inflammatory drug that also inhibits autophagy increasing lysosomal pH. The increase in pH inactivates lysosomal proteases, halting the progress of the autophagic process.
Smer 28 – positive mammalian autophagy regulator. Smer 28 increases autophagosome synthesis and clearance of model autophagy substrates (e.g. A53T α-synuclei).
SNX-2112 – ATP-competitve inhibitor of both Hsp90Alpha and Hsp90Beta heat shock protein. SNX-2112 induced autophagy in a time- and dose-dependent manner via Akt/mTOR/p70S6K inhibition. SNX-2112 induces significant apoptosis and exhibits anticancer properties, and is currently in clinical trials.
Spautin-1 – specific and potent autophagy inhibitor. Spautin-1 promotes the degradation of Vps34 PI3 kinase complexes by inhibiting two ubiquitin-specific proteases USP10 and USP13 which target the Beclin-1 subunit of Vps34 complexes. Deranges dengue virion maturation. Inhibits autophagy in a Beclin-1 independent manner in primary epithelial ovarian cancer cells. Spautin-1 selectively induces cell death of mutant p53-expressing cancer cell lines under confluency. Cell permeable.
STF-62247 – Specifically induces autophagic cell death in VHL (von Hippel-Lindau)-deficient renal carcinoma cells both in vitro (IC50=625 nM) and in vivo. Renal cell carcinoma cell lines
were radiosensitized by induction of autophagy following treatment with STF-62247. Cell permeable.
Temozolomide – DNA alkylating agent with antitumor and antiangiogenic activity. Induces autophagy in a variety of cell lines. Clinically useful anticancer agent. Cell permeable.
Wortmannin – steroidal metabolite belonging to the viridin group isolated from Penicillium wortmannii in 1957. Wortmannin exhibits broad spectrum antifungal activity together with antitumor activity and has been reported to possess anti-inflammatory activity. Wortmannin is a potent inhibitor of selected kinases, phosphoinositide 3-kinase and myosin light chain kinase. It is also capable of neutrophil and formyl-Met-Leu-Phe-mediated phospholipase D activation, inhibition of autophagy, potentiation the LPS-induced nitric oxide (NO) production and induction of in vivo Alzheimer-like hyperphosphorylation in tau.
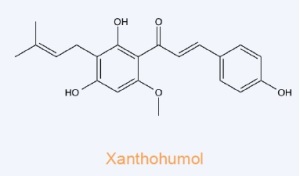 Xanthohumol – Binds to the N-terminal domain of valosin-containing protein (VCP), an essential protein for autophagosome maturation. Xanthohumol inhibits the function of VCP thereby impairing autophagosome maturation and resulting in accumulation of the microtubule-associated protein 1 light chain 3-II (LC3-II). Impairs prostate cancer cell growth and proliferation.
Xanthohumol – Binds to the N-terminal domain of valosin-containing protein (VCP), an essential protein for autophagosome maturation. Xanthohumol inhibits the function of VCP thereby impairing autophagosome maturation and resulting in accumulation of the microtubule-associated protein 1 light chain 3-II (LC3-II). Impairs prostate cancer cell growth and proliferation.
Cell permeable.
A tool to detect autophagy versus cytotoxicity
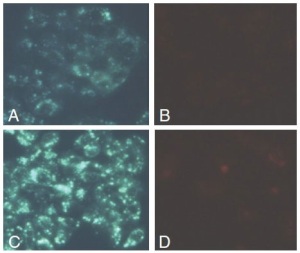
The Autophagy/Cytotoxicity Dual Staining Kit (Fig.3) is a fluorescence-labeled method for screening autophagy and cytotoxicity at the cellular level. The kit employs monodansylcadaverine (MDC), a fluorescent compound that is incorporated into multilamellar bodies by both an ion trapping mechanism and the interaction with membrane lipids, as a probe for detection of autophagic vacuoles in cultured cells. Propidium iodide is used as a marker of cell death. Tamoxifen, a known inducer of autophagy, is used as a positive control (and included in the kit).
Tools to measure ubiqutination
Usually the proteins to be degraded in autophagy are ubiquitinated and thus tagged for their fate. In a recent post I summarized our tools to measure ubiquitinated proteins – and I especially focused on Tandem Ubiquitin Binding Entities (TUBEs).
If you’re interested in detecting ubiquitinated proteins, take a few moments to read TUBEs – efficiently detect & purify poly-ubiquitinated proteins.
Share your experiences in autophagy and/or ubiquitination research, or ask any questions concerning the products and technologies described above through the form below!