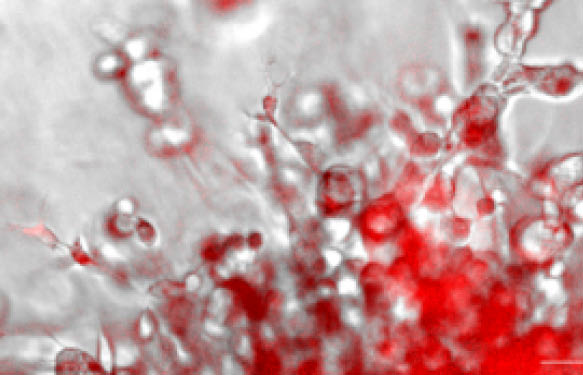
Recently, the non-FRET EnSens technology was launched for in vitro assessment of specific protease activities (Enzium, Inc.). Up to date, the EnSens substates were validated for the detection of protease activity in microplate assays. Now, Enzium has extended the applicability of their Ensens method to 3 D live-cell imaging. On the top of this, they announce that the experimental procedure is easy and non-toxic for cell cultures and co-cultures. So how does live-cell imaging help you locate protease activities in in vitro tumor invasion assays ?
The identification of small molecules able to modulate protease activity involved in cancers and metastasis continues to be a hot topic in drug discovery programs targeting especially these therapeutical targets. Over the last decades, several experimental methods have been developed to screen protease activities.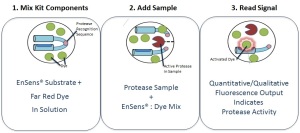
Recently, Enzium launched a non-FRET assay that enables unique (at the level of selectivity and versatility) protease measurements: the EnSens technology platform.
EnSens substrates for Protease Activity measurements in microplate assays
These EnSens Protease-specific substrates are modular “dumbbell” comprising two domains (dye-binding domain + blocking domain) separated by a linker which contains the highly specific protease recognition sequence. This linker brings the specificity of the EnSens protein substrate. When the linker is cleaved by the protease of interest, the dye-binding domain is free and activate a Far Red dye which in turn is generating a fluorescent signal.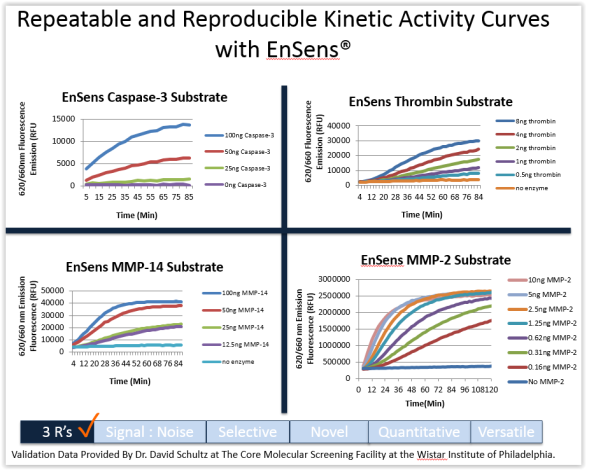
The EnSens technology surpass all limitations of FRET and FRET-like assays in plate reader fluorimetry applications.
Compared to these assays (in which the protease substrate is coupled to a fluorophore and a quencher), it has been demonstrated that the EnSens technology reaches unique detection levels by optimizing selectivity, robustness and accuracy of screening assays of diverse proteases (MMPs, ADAM10 and 17 (TACE) but also Furin, Thrombin and Factor Xa).
EnSens substrate to selectively assess Protease Activity in live-cell imaging and 3D tumor invasion cellular models
Recently, the EnSens technologyhas been proven to be compatible with 3D tumor invasion assays and fluorescent microscopy.
In a collaborative work, researchers from BioTek Instruments and Enzium have designed a smart and easy-to-use method to measure Protease activity and phenotypic effects of small molecules in tumoroid cell-based assays.
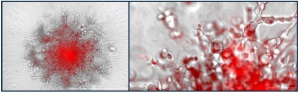
This new method is based on non-toxic, protease-selective substrates and far-red fluorescent dyes (the same as used in the classical biochemical EnSens assays described above) to be mixed directly into invasion matrix / scaffold / hydrogel (e.g. Matrigel®, or collagen gel), enabling real-time monitoring of specific protease activity by fluorescent microscopy in presence (or not) of bioactive small molecules (such as the hsp70 inhibitor Oridonin).
The protocol is ideal to monitor extracellular matrix enzyme activities in tumoroid cell models. During metastatic processes penetration of cancer cells through the basement membrane is a critical step in which MMPs especially MMP-14 play a critical role. MMP-14 is crucial for basement membrane and interstitial matrix degradation and helps cells gain the ability to penetrate the extracellular matrix.
Willing to track and monitor specific proteases such as MMP-14, MMP-2, MMP-3/MMP-10, MMP-9, MMP-13, MMP-25, ADAM10, ADAM17, Factor Xa, Furin and, Thrombin?
Interested in testing molecules in in vitro tumor invasion assays?
Do not hesitate to contact me with the form below.