As detailed in a previous post, rRNA depletion with kits such as Ribo-zero greatly increases the number of usable reads in RNASeq studies, but this one-size-fits-all approach is not appropriate for some researchers. In eurkaryotic studies, researchers often find that poly(A) enrichment using the BioMag® SelectaPure mRNA System making use of BioMag® Oligo (dT)20 Particles is a cost-effective means to separate mRNA from rRNA and tRNA. Prokaryotic mRNAs are not polyadenylated however, so poly(A) enrichment is not a possibility for researchers working with microorganisms. Although there exist rRNA depletion kits for bacteria, the organism-to-organism variability in rRNA sequences may lead to suboptimal depletion in some species and even the unintended depletion of specific mRNAs.
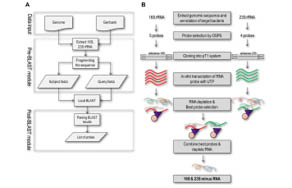
An article published in PLoS ONE (DOI: 10.1371/journal.pone.0074286) has described a computer program specifically for the purpose of designing rRNA depletion probes for various organisms. Using the example of the 16S and 23S rRNAs of Mycobacterium smegmatis, the authors report improved mRNA integrity and abundance using this approach compared to using MICROBExpress™.
The organism-specific probe selection software is available for download here. Once designed, researchers can purchase Biotin-TEG DNA oligonucleotides and BioMag® Nuclease-Free Streptavidin Particles for efficient depletion of rRNA from their organism of choice.
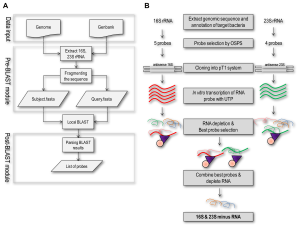
doi:10.1371/journal.pone.0074286.g001