We all react to external factors, even the most cold-blooded person. We might hide our emotions, but there they are.
The same happens in cancer. Cells (both the tumour cells and the normal ones) react to the environment in one direction or another. In this post, we will see how “kinome” analysis might help Researchers in better understanding cellular interactions in tumour microenvironment (TME).
In previous posts, we have put our spotlight on what is happening in the tumour microenvironment (TME), and how tumour cells, for example, avoid their destruction by evading the immune response from the normal cells.
This can be achieved either by secreting different molecules, by modifying glycosylation patterns, etc. This happens because the normal, healthy cells react to the stimulus provided by the tumour cells. Without “being aware” (if cells can be aware of something, which is another topic), normal cells cooperate to the expansion of the cancer, and this is done by the ability of cancer cells to modify the signaling pathways inside other cells.
There is a number of articles studying the signaling pathways relevant in different types of cancer. (1, 2) They combine mathematical and statistical models with experimental data.
However, we still lack a deeper knowledge. Increasing our understanding on how the cancer manages to survive and expand will allow to design more targeted therapies for the future. And this is key in the fight against cancer.
The kinome
Signaling can be done through a variety of enzymatic reactions, but one of the most relevant (and relatively known) is phosphorylation.
Study of the kinome is key to understanding the specific mechanisms, as we indicated previously, explaining how a cancer cell is able to modify its environment, thus paving the way for the design of better therapies, or helping to understand resistance to existing ones.
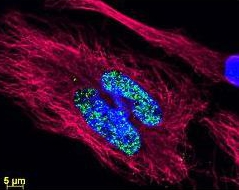
![Ezh 2 (phospho Thr487) antibody [EPR1410] (Cat. No. 210GTX62838).](https://beingbioreactive.files.wordpress.com/2099/09/pca_009.jpg)
We will discuss about them in this series of posts for TME and “kinome analysis”.
TME and biomarkers – when you already know something and want to know more
When you already know what is the pathway, or even the specific kinase, involved in the pathological condition that you are studying, then why go somewhere else?
This has been the “traditional” approach, and is still being used by a high number of laboratories, mainly doing Western Blot (WB), Immunofluorescence (IF) and/or Immunohistochemistry (IHC) studies. And it is still a very valuable source of information, both at the academic as well as at the clinical level (e.g. Pathological Anatomy laboratories).
Choosing well-validated antibodies here is key. The usual suspects include mTOR (phospho and non-phospho), Akt (with the different possible phosphorylations and isoforms), phospholipids, etc.
Different cancers will have different markers, as cancer is quite an heterogeneous group of diseases. For example, in prostate cancer, phosphorylation status of EZH2 and PTEN are key processes.
Studying phosphorylated vs. non-phosphorylated ratios
For a number of years, it is also possible to study, simultaneously, how phosphorylation affects a given biological process by studying the ratio between phosphorylated and non-phosphorylated forms of a kinase. There are several platforms in the market, but infra-red detection is quite popular.
When using this kind of platforms (e.g. Li-Cor Odyssey®), it is important to choose validated primary & secondary antibodies, optimised for this system. This way, we will optimise our experiments, allowing us to detect down to the picogram scale, with a higher signal-to-noise ratio. Infra-red detection can be used either in standard WB, or in order to perform in-cell western.
What about you?
A clear advantage is that traditional techniques (WB, IHC, IF) are easily accessible to most laboratories, and technical skills required for their performance are quite state-of-the-art (if I may insist, always that one uses validated antibodies). However, they are not suitable when either a large number of samples needs to be studied, or for quantitative results, or when one does not know what markers are relevant in a given experimental setting.
If you are in these cases (i.e. a large cohort or unknown markers), stay tuned or contact me!