Following our series of posts on the tumour microenvironment (TME), we will put our spotlight today on exosomes.
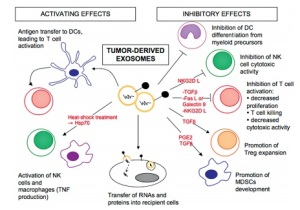
TME is composed on myofibroblasts, extracellular matrix and many other cell types. The tumour communicates with its microenvironment through cytokines, growth factors, chemokines, miRNAs, etc, as previously seen. Exosomes are nanosized extracellular vesicles (EVs) that allow communication between cells. They seem to play a role in the progression of some cancers (e.g. prostate cancer, glioblastoma), as well as in resistance to cancer therapies (1, 2). In fact, they modulate the immune response, explaining their role in either fighting the tumour or helping the tumour cells evade the immune system (3).
Exosomes contain a variety of factors to perform their role, including lipids, proteins, mRNA and miRNAs. Though most exosomes share the same surface markers, their content varies depending on cell type, explaining their different effects. Knowing the compounds present in exosomes can be useful not only to design new therapies, but also to find novel biomarkers for the diagnosis and prognosis of cancer (4).
Much research is still needed to know more about cancer-specific exosomes and their specific role in its progression and dissemination, but there are already some tools available to support this area of research. But first of all… what is an exosome?
What is an exosome?
Cells secrete several types of vesicles. The origin of some vesicles lies on the cell membrane. Other vesicles (EVs) are derived from the cytoplasm, and therefore, contain cytoplasmic factors (e.g. proteins, lipids, mRNA, miRNAs).

Depending on their size, these EVs can be divided into different subcategories. The smallest ones are called exosomes. Still, there is still no harmonization on the real definition of what an exosome is, so sometimes it is preferred to use the generic name of EVs (5). This difficulty in defining what a real exosome is, or whether an EV is an exosome or not is mainly caused by the lack of a harmonized method for exosome isolation.
Exosomes (or EVs in general) are isolated by ultracentrifugation methods. Some recent findings indicate that in fact, ultracentrifugation is not enough to separate exosomes from other types of EVs, and further isolation by sucrose gradients may be needed (6).
To make things worse, there is no consensus either on the markers that allow to differentiate exosomes from other EVs. There are some reviews to try to define a “canonical” exosome based on previous findings (7). In any case, there seems to be some consensus on what surface markers can be used to identify exosome-enriched subcellular fractions by flow cytometry or Western Blot. These include CD9, CD63, CD81, TSG101 and RAB27A, among others.
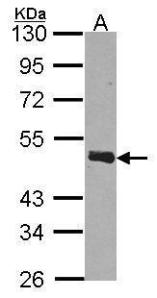
For identification of exosomes using antibodies, specially by flow cytometry, it is recommended that the epitope recognised by the antibody is in the area exposed to the environment, as epitopes located inside the exosomes will not be detected. In this sense, there are few antibodies which have been validated for detection of exosomal markers by flow cytometry, so using small, validation sizes, may be a good approach to try different antibodies and find the right one for your specific experimental model. We can also help on choosing the right antibody, as we have a wide feedback from researchers all over Europe!
So once you have your exosomes identified, what can you do with them? Stay tuned for our coming posts!
Working on exosomes and cancer? Or on exosomes and other diseases? Feel free to leave your comments!
References
1.- Zhao, H. et al. Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5-9; San Diego, CA. Philadelphia (PA): AACR; Cancer Res 2014;74(19 Suppl):Abstract nr 3594. doi:10.1158/1538-7445.AM2014-3594.
2.- Godlewski, J. et al. Neuro Oncology 2014 : nou292v1-nou292. doi: 10.1093/neuonc/nou292.
3.- Bobrie, A. et al. Traffic 2011; 12: 1659–1668. doi:10.1111/j.1600-0854.2011.01225.x.
4.- Challagundla, K.B. June 2014, Vol. 14, No. 5 , Pages 565-574. doi:10.1586/14737159.2014.922879.
5.- van der Pol, E. et al. Pharmacological Reviews July 2012 vol. 64 no. 3 676-705. doi: /10.1124/pr.112.005983.
6.- Bobrie, A. et al. J Extracell Vesicles. 2012; 1: 10.3402/jev.v1i0.18397. doi: 10.3402/jev.v1i0.18397.
7.- Théry, C. et al. Nature Reviews Immunology 9, 581-593. doi:10.1038/nri2567.