Cancer research is increasingly focusing on the tumour microenvironment (TME). Several studies have shown that tumours depend on external signals for maintenance and expansion. It is therefore needed to have a deeper knowledge of the cross-talk between tumour cells in the stroma (fibroblasts, adipocytes, endothelial cells and macrophages) and their microenvironment which also includes the study of interactions between cancer cells and cancer stem cells. TME studies also involve soluble factors, signaling molecules, extracellular matrix proteins (ECM) and other factors that help the tumour grow and invade other tissues, protect it from the host immune system, and contributes to therapeutic resistance in some cases (1). In a previous post, we discussed the role of Cox-2 signaling and PGE2 in TME. Also, we have already discussed the role of inflammation and the modification of the host’s immune response by cancer cells.
Today, we would like to focus on how TME affects the glycosylation of proteins involved in tumour progression.

Reports indicate that malignant transformation of cells is associated with aberrant glycosylation presented on the cell-surface (e.g. aberrant expression and glycosylation of mucins, abnormal branching on N-glycans, and increased presence of sialic acid on proteins and glycolipids). These changes in glycosylation influence cancer progression by affecting the capability of the tumour to disseminate and metastasise.
Aberrant glycans enable binding to the cells in the TME, through glycan-binding receptors (lectins such as siglecs, galectins and selectins). Some glycan alterations in carcinoma cells and their effect on lectin recognition have been described in the literature (2). All lectins facilitate interactions with the TME and therefore can effect cancer progression, meaning that they are now a focus to study the development of new therapies, including biologics (e.g. therapeutic mAbs).
The reality is, however, that mechanisms of glycosylation in cancer are still poorly understood. Selectin is the only lectin which mechanisms have been (at least partially) characterised. It has also been indicated that changes in glycosylation patterns in tumour cells are mainly mediated by an elevation of GlcNAc transferase-V activity, causing an increase in size and branching of the cell surface glycans, which in turns actively promotes differential recognition by galectins and selectins (3). Some other studies focus on the glycosylation pattern of exosomes rather than cells (4). More studies are needed in order to understand how glycosylation profiles affect cancer metastatic spread and resistance to therapy, as cancer-associated aberrant glycosylation has been identified in virtually every type of cancer.
Aberrations in glycosylation could be used as targets for diagnosis, prognosis or therapy. The ability to distinguish differences in the glycosylation of proteins between cancer and control patients (or responsive and non-responsive patients) would allow for the development of new diagnostic or prognostic biomarkers (5).
How to study glycosylation in cancer and TME?
Several publications use different approaches. Some publications study the glycosylation enzymatic pathways, and identify the nature and identity of the glycans in cancer cells or TME cells by MALDI MS (6). This can be cumbersome, as these techniques only allow for the characterisation of the different glycan chains. This is not time or cost efficient, as you cannot determine the glycosylation status of a therapy-resistant cancer vs a therapy-responsive one, for example.
Some other studies, however, rather than studying the glycosylation changes per se, study the regulation and localisation of lectins and how they relate to regulation of cancer cell adhesion and invasion (7). These studies are more specific, as they identify the lectin reacting to glycosylation changes, and therefore identify the rough glycan changes occurring in the cell/sample type (8). Specific techniques include lectin histochemical staining, lectin affinity chromatography, lectin-specific ELISA assays, as well as lectin arrays.
Solution #1 – Lectin-specific antibodies
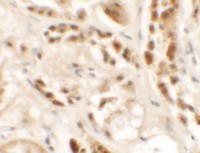
Some publications study lectin-mediated cancer progression by means of IHC detection of some relevant markers. The advantage of IHC is that not only presence is detected, but also the localisation. It is not a quantitative technique though, but can be used in Pathological Anatomy laboratories to follow progression of cancer. Siglec-15, for example, is expressed in Tumour-Associated Macrophages (TAM) in several cancer types.
Other relevant markers in the literature include Siglec-1, Siglec-2 (CD22), Siglec-9, Galectin-1, Galectin-3 and P-selectin.
Solution #2 – Lectin-specific ELISAs & assays
The utility of lectins has been clearly demonstrated by their ability to specially identify glycan changes to tumour cells or tumour produced proteins. Most common lectins involved in cancer progression include galectins, selectins, siglecs and others. Several ELISA kits are available for quantification of changes in lectin levels involved in glycosylation-mediated cancer progression: Galectin-1, Galectin-3, L-selectin, P-selectin, E-selectin and Siglec-9.
Solution #3 – Lectin-specific & glycosylation antibody arrays
Arrays are a sound approach to screen for glycosylation changes associated to tumour progression. Rather than going one marker by one (as in the previous solutions), multiplexed arrays allow for the profiling of glycosylation changes of a sample or pool of samples, by measuring the levels of 40 lectins simultaneously. Therefore, arrays allow the researcher to identify and profile the glycans in tumour samples, determine whether the biomarker of interest has glycan moieties and/or find specific glycan binding ligands in biological samples.
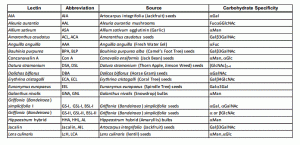
One approach is to measure global glycosylation levels with a lectin based array. This array allows for simultaneous measurements of the levels of 40 lectins which bind specifically to certain glycosylation patterns. Therefore, lectin arrays allow, for the semi-quantitative analysis of lectin-glycoprotein interactions present in any sample. For example, measuring the glycosylation patterns between a primary and a cancer cell’s conditioned media may elucidate potential changes in the overall glycosylation patterns in these cell’s secretome.
There are new solutions which are being developed now, including arrays contaning up to 60 lectins, as well as new applications for existing and customised arrays for specific protein and glycosylation detection using glycosylation arrays. So stay tuned!
Do you have any questions? Just leave your comments or remarks below!
References
1.- Melody A. Swartz, Noriho Iida, Edward W. Roberts, et al. Cancer Res May 15, 2012 72; 2473. doi: 10.1158/0008-5472.CAN-12-0122.
2.- Irina Häuselmann and Lubor Borsig. Front Oncol. 2014; 4: 28. doi: 10.3389/fonc.2014.00028 PMCID: PMC3923139.
3.- Varki A, Kannagi R, Toole BP. Glycosylation Changes in Cancer. In: Varki A, Cummings RD, Esko JD, et al., editors. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 44. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1963/.
4.- Catarina Roma-Rodrigues, Alexandra R. Fernandes, and Pedro Viana Baptista, “Exosome in Tumour Microenvironment: Overview of the Crosstalk between Normal and Cancer Cells,” BioMed Research International, vol. 2014, Article ID 179486, 10 pages, 2014. doi:10.1155/2014/179486.
5.- Franca Maria Tuccillo, Annamaria de Laurentiis, Camillo Palmieri, et al., “Aberrant Glycosylation as Biomarker for Cancer: Focus on CD43,” BioMed Research International, vol. 2014, Article ID 742831, 13 pages, 2014. doi:10.1155/2014/742831.
6.- Hugo Osório and Celso A. Reis. Mass Spectrometry Data Analysis in Proteomics. Methods in Molecular Biology Volume 1007, 2013, pp 301-316. doi 10.1007/978-1-62703-392-3_13.
7.- Bassagañas S, Carvalho S, Dias AM, Pérez-Garay M, Ortiz MR, et al. (2014) Pancreatic Cancer Cell Glycosylation Regulates Cell Adhesion and Invasion through the Modulation of a2ß1 Integrin and E-Cadherin Function. PLoS ONE 9(5): e98595. doi:10.1371/journal.pone.0098595.
8.- Christiansen, M. N., Chik, J., Lee, L., Anugraham, M., Abrahams, J. L. and Packer, N. H. (2014), Cell surface protein glycosylation in cancer. Proteomics, 14: 525–546. doi: 10.1002/pmic.201300387.