

Download the Application Note
Explore how to optimize Lipid Nanoparticules for mRNA delivery for drug development.
This study highlights key formulation parameters: buffer composition, lipid selection; crucial for enhancing transfection and therapeutic efficacy.
Introduction
With the COVID-19 pandemic and the development of mRNA vaccine, LNPs gained notoriety in the field of nucleic acid - based drug delivery [1]. Once encapsulated into these vesicles, nucleic acid are protected from enzymatic degradation and safely delivered to the cytosol, representing the most advanced non-viral nucleic acid delivery system in clinical applications [2].
Four lipids represent the main components of LNPs: 1) phospholipids and 2) cholesterol – necessary for particle formation and stability, 3) ionizable lipids - allowing the binding to negatively charged nucleic acid and 4) PEGylated lipids – enhancers of stability and circulation time within the biological system (Figure 1) [3]. These components, similar to the cellular membrane lipids, allow the fusion of LNPs with cell membranes and the release of nucleic acids in the cytosol.
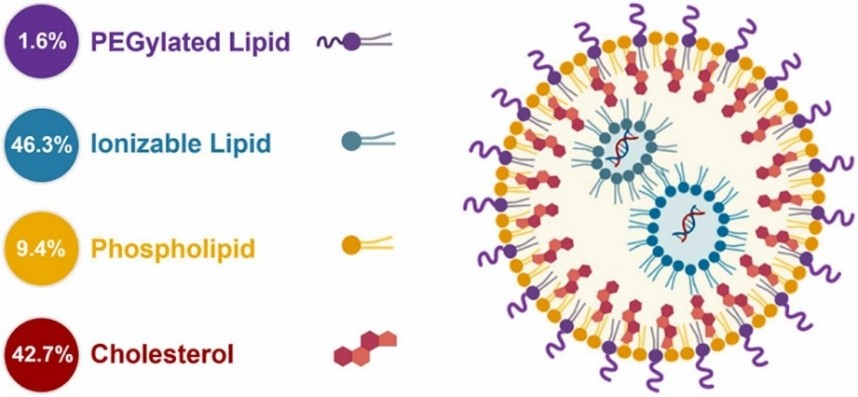
Figure 1 - Components of LNPs - Lipids are color coded and the molar percent lipid composition values are provided for the Comirnaty™ vaccine (COVID-19 from Pfizer & BioNTech) [4].
Several factors influence the efficiency of LNPs, including particle size, which affects stability, biodistribution, and cellular uptake. Smaller LNPs (<100 nm) exhibit enhanced cellular uptake and prolonged circulation time contrary to larger LNPs (>100 nm). Another key factor is the LNP surface charge influencing stability: the stronger the surface charge, the stronger the stability [3].
At Tebubio, we offer an extensive library of lipids for LNP production, as well as ready-to-use LNP formulations. As part of the development of our new mRNA delivery platform, we tested and optimized various lipid formulations coming from our suppliers and partners. In this study, we go through our step process to encapsulate your mRNA with various formulations and also their potencies in two different cell lines: the “classical” HEK293 and the HCT116 cell line, one of the model of colon cancer. In our lab, we have tested different formulations using lipids from our different suppliers, including Echelon, Polyplus, ABP Biosciences and Certest to increase the encapsulation and cellular transfection efficiency. The aim of this study is to describe these different steps using an mRNA coding the eGFP protein, encapsulated in different LNP formulations from different suppliers and transfected in two different cell lines, providing the ability to discriminate the transfection and expression differences in regards to LNP properties and as well as the cellular model.
Materials & Methods
mRNA synthesis
The plasmid was synthesized and amplified by GenScript. Next, 50 μg of plasmid was digested with BspQI (NEB, #R0712). The DNA was purified with the NuceloSpin gel and PCR clean-up kit (Macherey Nagel, #740609) according to the manufacturer’s instructions.
IVT
The RNA was synthesized with the T7-FlashScribe Transcription Kit (CellScript, #C-ASF3507) according to the manufacturer’s instructions, in the presence of N1-Methyl-pseudouridine-5'-Triphosphate (Trilink, #N-1081) replacing UTP. Then, a treatment with the RNase-free DNAse I from the T7 mScript Standard mRNA Production System was performed according to the manufacturer’s instructions. Finally, the RNA was purified using Mag-Bind® TotalPure NGS (Omega Biotek, #M1378-01) on Hamilton ML Prep Robot according to the manufacturer’s instructions.
Capping
The capping of the RNA was performed using the ScriptCap m7G Capping System and ScriptCap 2'-O-Methyltransferase Kit from (CellScript, #C-SCCE0625 and #C-SCMT0625) according to the manufacturer’s instructions and purified with Mag-Bind® TotalPure NGS (Omega Biotek, #M1378-01) on Hamilton ML Prep Robot. The plasmid quality and concentration was verified by NanoVue Plus (GE Healthcare) and TapeStation 4150 (Agilent). Endotoxin level were measured by Limulus Amebocyte Lysate Endosafe®-PTS Cartridges (LAL) (# PTS2005F), Endosafe®-Portable Test System (Charles River Laboratories).
mRNA-LNP formulation and chracterization
The mRNA of interest coding eGFP was diluted in buffer A (Sodium acetate pH 4.8, 50 mM final) or in standard commercial buffer to achieve a final mRNA concentration between 0.1-0.5 mg/ml, depending on the N/P ratio desired.
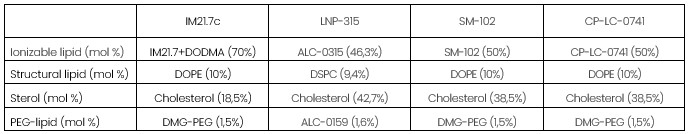
The following mixes have been prepared, off note, LNP-315 is an off-the-shelf LNP preparation (ABP Biosciences) ready to use for mRNA encapsulation:
Prior to formulation, lipids were heated at 55°C for 5 minutes, to dissolve eventual cholesterol aggregates, then mixed using the Nanoassemblr Spark nanoparticle formulation system (Precision NaNosystems, Cytiva), and NanoAssemblr® Spark® cartridges (NIS0013, Precision NaNosystems, Cytiva) according to the manufacturer’s instructions with the following conditions: Flow rate ratios of 3, N/P between 4-8 (mRNA between 5-10 μg). Formulated LNPs were then diluted 1:1 in PBS buffer (depleted in Ca2+ and Mg2+) and Dynamic light scattering (DLS) analyses are performed after a 1:10 dilution of LNPs in PBS , with the Zetasizer ultra red (Zetasizer®, Malvern Panalytical) using Cumulants analysis. For encapsulation efficiency measurements, Quant-iT RiboGreen RNA Assay Kit (ThermoFisher, #R11490) is used, following manufacturer’s instructions.
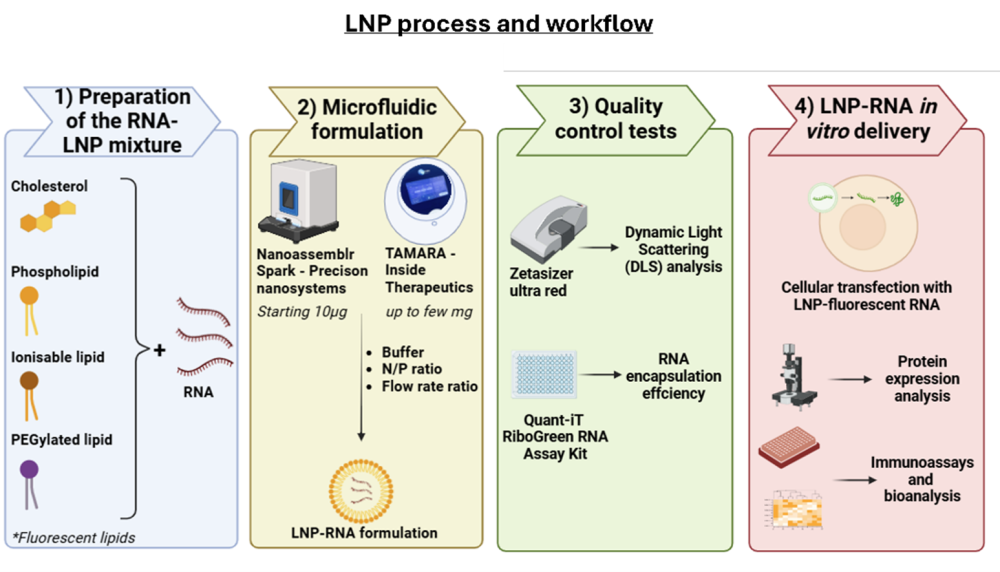
Figure 2 – LNP process and workflow at Tebubio - The first step consists of preparing the RNA and LNP mixtures for the encapsulation using the Nanoassemblr Spark from Precision nanosystems (for small quantities – starting 10 μg) or TAMARA from Inside Therapeutics (allowing a scale up – up to few mg) . The quality of the resulting formulation will be evaluated by Dynamic Light Scattering (DLS) analysis and RNA encapsulation efficiency. These RNA-LNP formulation are then tested in vitro after cell lines transfection, evaluating the protein expression and its effect by performing immunoassays and bioanalysis studies.
In vitro cell culture
Human embryonic kidney cells (HEK293) and human colorectal carcinoma cancer cell line (HCT116) were seeded in a T-75 flask with the corresponding medium at 37 ̊ C with 5% CO2. After reaching 80% confluency, cells were trypsinated and cultured in a 96-well plate with a seeding density of 20 000 cell per well.
mRNA delivery via LNP-based transfection
Different LNP mixtures with encapsulated mRNA-eGFP quantities ranging between 40-250 ng were 10x diluted in the appropriate cell medium and incubated with the cells. As a positive control, mRNA-eGFP was transfected using the mRNA-Fect reagent (RJH BioSciences, #SKU 80-40). Images were acquired using fluorescence microscopy (Ti:2, Nikon) 24 hours after transfection.
Results
Below are some of the key points we want to highlight regarding mRNA formulation:
mRNA preparation
mRNA preparation is crucial, starting from its sequence design (UTRs, codons, etc) up to its actual synthesis (cap1, polyA, modified UTPs) in order for the mRNA to be efficiently translated in vivo (or in vitro). Encapsulation is less reliant on these parameters but are influenced by others, one of them being the pH at which the encapsulation is performed.
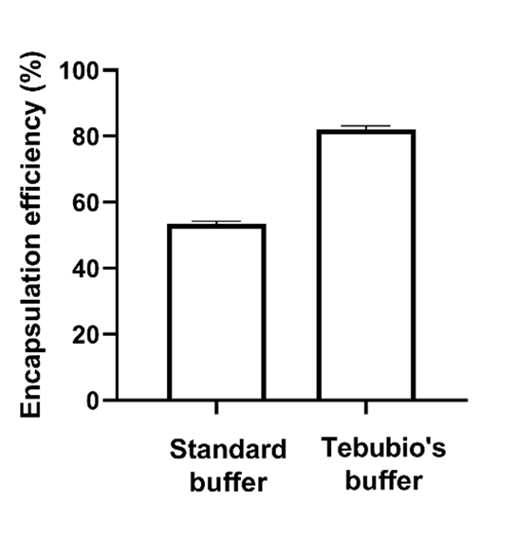
Figure 3 - Encapsulation efficiencies of eGFP-mRNA formulated in LNP-0315 (ABP-Biosciences) using a commercially available buffer at pH 5.5 and a buffer prepared in our laboratory (pH 4.8)
LNP formulation relies on the interaction of mRNA with the ionizable lipid. The efficiency of this interaction is dictated by the relationship between the pKa of the ionizable lipid and the pH of the mRNA solution, the latter must be much lower than the former. Figure 3 highlights the importance of optimizing the mRNA solution buffer in order to achieve proper encapsulation of the mRNA. In that experiment, we compared the encapsulation efficiencies of eGFP-mRNA in LNP-0315 following the use of a “standard” buffer (pH 5.5) compared to a buffer prepared internally (pH 4.8). From this result, it appears clearly that the “standard” buffer, even though below the pKa of the ALC-0315 lipid (pH 6.09), led to subpar encapsulation of the mRNA. In contrast, decreasing the pH up to 4.8 (or lower, data not shown) led to significant increase of the encapsulation efficiency.
This simple experiment really shows the importance of knowing the overall system one work with before preparing LNPs.
Choice of ionizable lipids
The second important parameter we want to emphasize is the choice of ionizable lipids. This choice is driven by the application you’re interested in (tropism of the ionizable lipid, downstream applications, etc).
To illustrate this we present data from 4 different formulations containing different ionizable lipids.
Without any formulation optimization, we obtained particles of good quality (PI< 0.2, except for SM-102 where PI = 0.26, data not shown). Similarly, encapsulation efficiency was > 80 % for all the formulation tested, except for SM-102 where encapsulation efficiency was only at ~65 %). This result suggest that formulation optimization would be needed for this particular LNPs.
We then further characterized the 4 different formulations regarding protein translation (eGFP) in 2 different cellular models: HEK293 cells (Figure 4) or colon cancer cell model HCT116 (Figure 5).
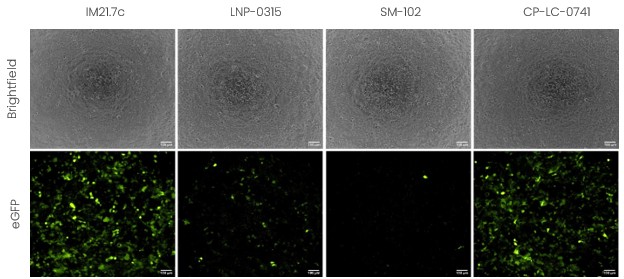
Figure 4 - Evaluation of the encapsulation efficiency and in vitro transfection of LNP-mRNA in HEK293 cells using formulations with different ionizable lipids.
The results obtained in HEK cells clearly show the importance of selecting proper ionizable lipids for your application. From these data, it appears clearly that the formulations using IM21.7C (Polyplus – Sartorius) and CPLC-0741 (Certest) led to higher transfection efficiency and protein expression level compared to the other 2 formulations tested here.
On the other hand, when looking at the same formulation in HCT116 cells, we observe a different behavior (Figure 5). Indeed, in this cell line, all formulations led to very high transfection efficiency and good protein expression.
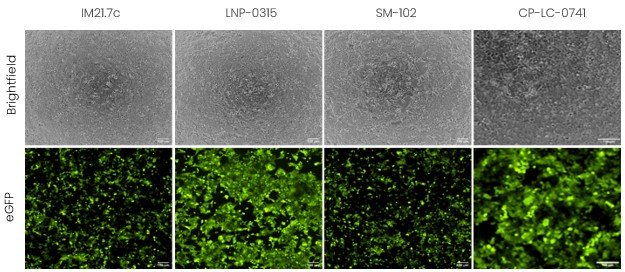
Figure 5 - Evaluation of the encapsulation efficiency and in vitro transfection of LNP-mRNA in HCT116 cells using formulations with different ionizable lipids.
From these data, the formulations that seems the most promising for future studies would be LNP-0315 (ABP-BioSciences) and CP-LC-0741. The emphasis here is that one lipid does not fit all, but a careful selection and formulation screening has to be conducted in order to find the solution that works best for the desired application.
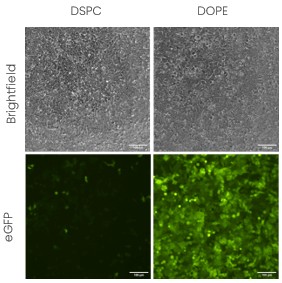
Figure 6 - Evaluation of the impact of structural lipids in transfection efficiency in HEK cells.
To further strengthen this idea, we present another set of in vitro transfection in HEK293 cells (Figure 6) using the same formulation (same ionizable lipid and lipid proportions) except that we changed the structural lipid from DSPC to DOPE. All the other parameters and lipid proportions were kept identical. The impact of DOPE on in vitro endocytosis being already well documented, this result is coherent with the literature [5]. However, it has also been shown that DSPC, the structural lipid found in Pfizer and Moderna vaccines, lead to more stable formulation [5] which is also a crucial parameters owing to the labile nature of LNPs. This exemplifies that formulation design is key, even in vitro, and that formulation screening remains essential when doing discovery work.
Conclusion
This study focuses on demonstrating the importance of LNP formulation optimization, from very basic, and well known, factors such as working solution pH to more fine tuning of the LNP formulation (type of ionizable lipid or structural/helper lipid).
LNP formulation is a complex process influenced by multiple factors, including the buffer composition, mRNA concentration, and lipid composition. However, it remains a crucial step in the development of RNA-based therapeutics.
Moreover, the cellular uptake and delivery of mRNA-LNPs involve a series of intricate steps, starting with LNP attachment to the cell surface, internalization via endocytosis, endosomal escape into the cytosol, and finally, mRNA translation by ribosomes for protein expression. The complexity of this delivery process is further rendered by additional variables intrinsic to both the LNPs (e.g., shape and surface chemistry) and the cell surface (e.g., elasticity, receptor concentration, and diffusivity), making these interactions highly heterogeneous [6]. As a result, cell uptake kinetics and transfection efficiency can vary significantly depending on both the LNP composition and the cell type, as observed in our cellular transfection assays.
Our findings underscore the necessity of optimizing multiple parameters for vaccine development and highlight the importance of selecting an appropriate cellular model to accurately translate and predict in vivo outcomes. At Tebubio, we help you develop tailored LNP formulations to suit your drug of interest and desired cellular model.
Additionally, we offer comprehensive immunoassays and bioanalysis studies to evaluate delivery efficiency and therapeutic efficacy.
Realised with the support of:
![]()

-
Authors
Elise Abboud, PhD – Xavier Warnet, PhD
-
Affiliation
Tebubio - 39 rue de Houdan, Le Perray en Yvelines, France
-
References
[1] Gambaro R, Rivero Berti I, Limeres MJ, Huck-Iriart C, Svensson M, Fraude S, et al. Optimizing mRNA-Loaded Lipid Nanoparticles as a Potential Tool for Protein-Replacement Therapy. Pharmaceutics. juin 2024;16(6):771.
[2] Hald Albertsen C, Kulkarni JA, Witzigmann D, Lind M, Petersson K, Simonsen JB. The role of lipid components in lipid nanoparticles for vaccines and gene therapy. Adv Drug Deliv Rev. sept 2022;188:114416.
[3] Mehta M, Bui TA, Yang X, Aksoy Y, Goldys EM, Deng W. Lipid-Based Nanoparticles for Drug/Gene Delivery: An Overview of the Production Techniques and Difficulties Encountered in Their Industrial Development. ACS Mater Au. 8 nov 2023;3(6):600‑19.
[4] Fekete S, Doneanu C, Addepalli B, Gaye M, Nguyen J, Alden B, et al. Challenges and emerging trends in liquid chromatography-based analyses of mRNA pharmaceuticals. J Pharm Biomed Anal. 5 févr 2023;224:115174.
[5] Barbieri BD, Peeler DJ, Samnuan K, Day S, Hu K, Sallah HJ, et al. The role of helper lipids in optimising nanoparticle formulations of self-amplifying RNA. J Controlled Release. 1 oct 2024;374:280‑92.
[6] Patel N, Davis Z, Hofmann C, Vlasak J, Loughney JW, DePhillips P, et al. Development and Characterization of an In Vitro Cell-Based Assay to Predict Potency of mRNA–LNP-Based Vaccines. Vaccines. 10 juill 2023;11(7):1224.
Need Laboratory Services? Get a Personalized Quote Today!
Leverage our expert laboratory services for your RNA production needs. Contact us for a tailored solution and quotation.

